Switch to List View
Image and Video Gallery
This is a searchable collection of scientific photos, illustrations, and videos. The images and videos in this gallery are licensed under Creative Commons Attribution Non-Commercial ShareAlike 3.0. This license lets you remix, tweak, and build upon this work non-commercially, as long as you credit and license your new creations under identical terms.
6557: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
6557: Floral pattern in a mixture of two bacterial species, Acinetobacter baylyi and Escherichia coli, grown on a semi-solid agar for 24 hours
Floral pattern emerging as two bacterial species, motile Acinetobacter baylyi and non-motile Escherichia coli (green), are grown together for 24 hours on 0.75% agar surface from a small inoculum in the center of a Petri dish.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6550 for a video of this process.
See 6553 for a photo of this process at 48 hours on 1% agar surface.
See 6555 for another photo of this process at 48 hours on 1% agar surface.
See 6556 for a photo of this process at 72 hours on 0.5% agar surface.
See 6550 for a video of this process.
L. Xiong et al, eLife 2020;9: e48885
View Media

3606: Flower-forming cells in a small plant related to cabbage (Arabidopsis)
3606: Flower-forming cells in a small plant related to cabbage (Arabidopsis)
In plants, as in animals, stem cells can transform into a variety of different cell types. The stem cells at the growing tip of this Arabidopsis plant will soon become flowers. Arabidopsis is frequently studied by cellular and molecular biologists because it grows rapidly (its entire life cycle is only 6 weeks), produces lots of seeds, and has a genome that is easy to manipulate.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
This image was part of the Life: Magnified exhibit that ran from June 3, 2014, to January 21, 2015, at Dulles International Airport.
Arun Sampathkumar and Elliot Meyerowitz, California Institute of Technology
View Media

3424: White Poppy
3424: White Poppy
A white poppy. View cropped image of a poppy here 3423.
Judy Coyle, Donald Danforth Plant Science Center
View Media

1010: Lily mitosis 10
1010: Lily mitosis 10
A light microscope image of a cell from the endosperm of an African globe lily (Scadoxus katherinae). This is one frame of a time-lapse sequence that shows cell division in action. The lily is considered a good organism for studying cell division because its chromosomes are much thicker and easier to see than human ones. Staining shows microtubules in red and chromosomes in blue. Here, condensed chromosomes are clearly visible and are separating to form the cores of two new cells.
Related to images 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Related to images 1011, 1012, 1013, 1014, 1015, 1016, 1017, 1018, 1019, and 1021.
Andrew S. Bajer, University of Oregon, Eugene
View Media

3509: Neuron with labeled synapses
3509: Neuron with labeled synapses
In this image, recombinant probes known as FingRs (Fibronectin Intrabodies Generated by mRNA display) were expressed in a cortical neuron, where they attached fluorescent proteins to either PSD95 (green) or Gephyrin (red). PSD-95 is a marker for synaptic strength at excitatory postsynaptic sites, and Gephyrin plays a similar role at inhibitory postsynaptic sites. Thus, using FingRs it is possible to obtain a map of synaptic connections onto a particular neuron in a living cell in real time.
Don Arnold and Richard Roberts, University of Southern California.
View Media

6769: Culex quinquefasciatus mosquito larva
6769: Culex quinquefasciatus mosquito larva
A mosquito larva with genes edited by CRISPR. The red-orange glow is a fluorescent protein used to track the edits. This species of mosquito, Culex quinquefasciatus, can transmit West Nile virus, Japanese encephalitis virus, and avian malaria, among other diseases. The researchers who took this image developed a gene-editing toolkit for Culex quinquefasciatus that could ultimately help stop the mosquitoes from spreading pathogens. The work is described in the Nature Communications paper "Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes" by Feng et al. Related to image 6770 and video 6771.
Valentino Gantz, University of California, San Diego.
View Media

7012: Adult Hawaiian bobtail squid burying in the sand
7012: Adult Hawaiian bobtail squid burying in the sand
Each morning, the nocturnal Hawaiian bobtail squid, Euprymna scolopes, hides from predators by digging into the sand. At dusk, it leaves the sand again to hunt.
Related to image 7010 and 7011.
Related to image 7010 and 7011.
Margaret J. McFall-Ngai, Carnegie Institution for Science/California Institute of Technology, and Edward G. Ruby, California Institute of Technology.
View Media

3362: Sphingolipid S1P1 receptor
3362: Sphingolipid S1P1 receptor
The receptor is shown bound to an antagonist, ML056.
Raymond Stevens, The Scripps Research Institute
View Media

3395: NCMIR mouse tail
3395: NCMIR mouse tail
Stained cross section of a mouse tail.
Tom Deerinck, National Center for Microscopy and Imaging Research (NCMIR)
View Media

6803: Staphylococcus aureus aggregates on microstructured titanium surface
6803: Staphylococcus aureus aggregates on microstructured titanium surface
Groups of Staphylococcus aureus bacteria (blue) attached to a microstructured titanium surface (green) that mimics an orthopedic implant used in joint replacement. The attachment of pre-formed groups of bacteria may lead to infections because the groups can tolerate antibiotics and evade the immune system. This image was captured using a scanning electron microscope.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6804 and video 6805.
More information on the research that produced this image can be found in the Antibiotics paper "Free-floating aggregate and single-cell-initiated biofilms of Staphylococcus aureus" by Gupta et al.
Related to image 6804 and video 6805.
Paul Stoodley, The Ohio State University.
View Media

2322: Modeling disease spread
2322: Modeling disease spread
What looks like a Native American dream catcher is really a network of social interactions within a community. The red dots along the inner and outer circles represent people, while the different colored lines represent direct contact between them. All connections originate from four individuals near the center of the graph. Modeling social networks can help researchers understand how diseases spread.
Stephen Eubank, University of Virginia Biocomplexity Institute (formerly Virginia Bioinformatics Institute)
View Media

2520: Bond types (with labels)
2520: Bond types (with labels)
Ionic and covalent bonds hold molecules, like sodium chloride and chlorine gas, together. Hydrogen bonds among molecules, notably involving water, also play an important role in biology. See image 2519 for an unlabeled version of this illustration. Featured in The Chemistry of Health.
Crabtree + Company
View Media

6932: Axolotl
6932: Axolotl
An axolotl—a type of salamander—that has been genetically modified so that its developing nervous system glows purple and its Schwann cell nuclei appear light blue. Schwann cells insulate and provide nutrients to peripheral nerve cells. Researchers often study axolotls for their extensive regenerative abilities. They can regrow tails, limbs, spinal cords, brains, and more. The researcher who took this image focuses on the role of the peripheral nervous system during limb regeneration.
This image was captured using a stereo microscope.
Related to images 6927 and 6928.
This image was captured using a stereo microscope.
Related to images 6927 and 6928.
Prayag Murawala, MDI Biological Laboratory and Hannover Medical School.
View Media

2555: RNA strand (with labels)
2555: RNA strand (with labels)
Ribonucleic acid (RNA) has a sugar-phosphate backbone and the bases adenine (A), cytosine (C), guanine (G), and uracil (U). Featured in The New Genetics.
See image 2554 for an unlabeled version of this illustration.
See image 2554 for an unlabeled version of this illustration.
Crabtree + Company
View Media
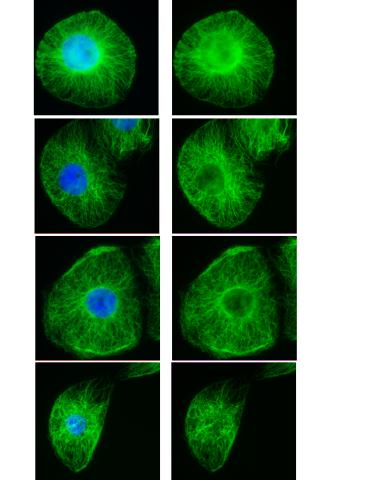
3443: Interphase in Xenopus frog cells
3443: Interphase in Xenopus frog cells
These images show frog cells in interphase. The cells are Xenopus XL177 cells, which are derived from tadpole epithelial cells. The microtubules are green and the chromosomes are blue. Related to 3442.
Claire Walczak, who took them while working as a postdoc in the laboratory of Timothy Mitchison.
View Media

2443: Mapping human genetic variation
2443: Mapping human genetic variation
This map paints a colorful portrait of human genetic variation around the world. Researchers analyzed the DNA of 485 people and tinted the genetic types in different colors to produce one of the most detailed maps of its kind ever made. The map shows that genetic variation decreases with increasing distance from Africa, which supports the idea that humans originated in Africa, spread to the Middle East, then to Asia and Europe, and finally to the Americas. The data also offers a rich resource that scientists could use to pinpoint the genetic basis of diseases prevalent in diverse populations. Featured in the March 19, 2008, issue of Biomedical Beat.
Noah Rosenberg and Martin Soave, University of Michigan
View Media

3497: Wound healing in process
3497: Wound healing in process
Wound healing requires the action of stem cells. In mice that lack the Sept2/ARTS gene, stem cells involved in wound healing live longer and wounds heal faster and more thoroughly than in normal mice. This confocal microscopy image from a mouse lacking the Sept2/ARTS gene shows a tail wound in the process of healing. See more information in the article in Science.
Related to images 3498 and 3500.
Related to images 3498 and 3500.
Hermann Steller, Rockefeller University
View Media

2339: Protein from Arabidopsis thaliana
2339: Protein from Arabidopsis thaliana
NMR solution structure of a plant protein that may function in host defense. This protein was expressed in a convenient and efficient wheat germ cell-free system. Featured as the June 2007 Protein Structure Initiative Structure of the Month.
Center for Eukaryotic Structural Genomics
View Media

3723: Fluorescent microscopy of kidney tissue
3723: Fluorescent microscopy of kidney tissue
Serum albumin (SA) is the most abundant protein in the blood plasma of mammals. SA has a characteristic heart-shape structure and is a highly versatile protein. It helps maintain normal water levels in our tissues and carries almost half of all calcium ions in human blood. SA also transports some hormones, nutrients and metals throughout the bloodstream. Despite being very similar to our own SA, those from other animals can cause some mild allergies in people. Therefore, some scientists study SAs from humans and other mammals to learn more about what subtle structural or other differences cause immune responses in the body.
Related to entries 3725 and 3675.
Related to entries 3725 and 3675.
Tom Deerinck , National Center for Microscopy and Imaging Research
View Media

6766: Ribbon diagram of a cefotaxime-CCD-1 complex
6766: Ribbon diagram of a cefotaxime-CCD-1 complex
CCD-1 is an enzyme produced by the bacterium Clostridioides difficile that helps it resist antibiotics. Using X-ray crystallography, researchers determined the structure of a CCD-1 molecule and a molecule of the antibiotic cefotaxime bound together. The structure revealed that CCD-1 provides extensive hydrogen bonding and stabilization of the antibiotic in the active site, leading to efficient degradation of the antibiotic.
Related to images 6764, 6765, and 6767.
Related to images 6764, 6765, and 6767.
Keith Hodgson, Stanford University.
View Media

3477: HIV Capsid
3477: HIV Capsid
This image is a computer-generated model of the approximately 4.2 million atoms of the HIV capsid, the shell that contains the virus' genetic material. Scientists determined the exact structure of the capsid and the proteins that it's made of using a variety of imaging techniques and analyses. They then entered these data into a supercomputer that produced the atomic-level image of the capsid. This structural information could be used for developing drugs that target the capsid, possibly leading to more effective therapies. Related to image 6601.
Juan R. Perilla and the Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign
View Media
2792: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 03
2792: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 03
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
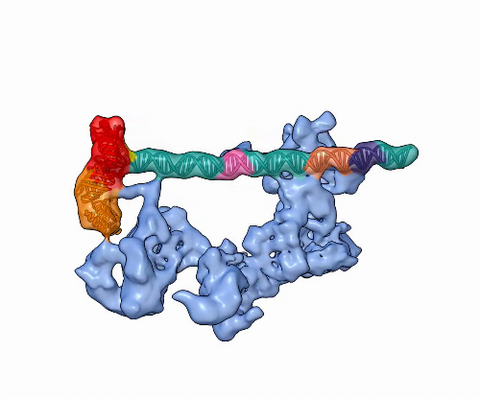
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
5730: Dynamic cryo-EM model of the human transcription preinitiation complex
Gene transcription is a process by which information encoded in DNA is transcribed into RNA. It's essential for all life and requires the activity of proteins, called transcription factors, that detect where in a DNA strand transcription should start. In eukaryotes (i.e., those that have a nucleus and mitochondria), a protein complex comprising 14 different proteins is responsible for sniffing out transcription start sites and starting the process. This complex represents the core machinery to which an enzyme, named RNA polymerase, can bind to and read the DNA and transcribe it to RNA. Scientists have used cryo-electron microscopy (cryo-EM) to visualize the TFIID-RNA polymerase-DNA complex in unprecedented detail. This animation shows the different TFIID components as they contact DNA and recruit the RNA polymerase for gene transcription.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab.
Related to image 3766.
Eva Nogales, Berkeley Lab
View Media

3766: TFIID complex binds DNA to start gene transcription
3766: TFIID complex binds DNA to start gene transcription
Gene transcription is a process by which the genetic information encoded in DNA is transcribed into RNA. It's essential for all life and requires the activity of proteins, called transcription factors, that detect where in a DNA strand transcription should start. In eukaryotes (i.e., those that have a nucleus and mitochondria), a protein complex comprising 14 different proteins is responsible for sniffing out transcription start sites and starting the process. This complex, called TFIID, represents the core machinery to which an enzyme, named RNA polymerase, can bind to and read the DNA and transcribe it to RNA. Scientists have used cryo-electron microscopy (cryo-EM) to visualize the TFIID-RNA polymerase-DNA complex in unprecedented detail. In this illustration, TFIID (blue) contacts the DNA and recruits the RNA polymerase (gray) for gene transcription. The start of the transcribed gene is shown with a flash of light. To learn more about the research that has shed new light on gene transcription, see this news release from Berkeley Lab. Related to video 5730.
Eva Nogales, Berkeley Lab
View Media
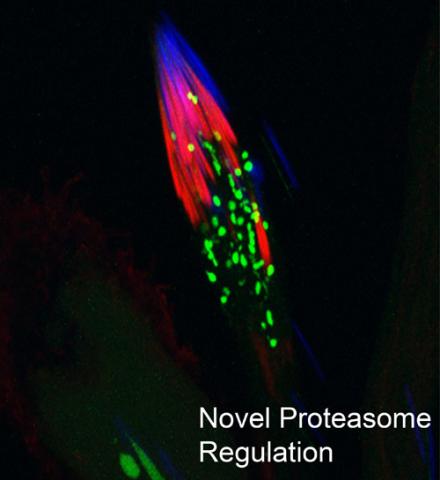
3451: Proteasome
3451: Proteasome
This fruit fly spermatid recycles various molecules, including malformed or damaged proteins. Actin filaments (red) in the cell draw unwanted proteins toward a barrel-shaped structure called the proteasome (green clusters), which degrades the molecules into their basic parts for re-use.
Sigi Benjamin-Hong, Rockefeller University
View Media

2392: Sheep hemoglobin crystal
2392: Sheep hemoglobin crystal
A crystal of sheep hemoglobin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media
2437: Hydra 01
2437: Hydra 01
Hydra magnipapillata is an invertebrate animal used as a model organism to study developmental questions, for example the formation of the body axis.
Hiroshi Shimizu, National Institute of Genetics in Mishima, Japan
View Media

2561: Histones in chromatin (with labels)
2561: Histones in chromatin (with labels)
Histone proteins loop together with double-stranded DNA to form a structure that resembles beads on a string. See image 2560 for an unlabeled version of this illustration. Featured in The New Genetics.
Crabtree + Company
View Media

3758: Dengue virus membrane protein structure
3758: Dengue virus membrane protein structure
Dengue virus is a mosquito-borne illness that infects millions of people in the tropics and subtropics each year. Like many viruses, dengue is enclosed by a protective membrane. The proteins that span this membrane play an important role in the life cycle of the virus. Scientists used cryo-EM to determine the structure of a dengue virus at a 3.5-angstrom resolution to reveal how the membrane proteins undergo major structural changes as the virus matures and infects a host. The image shows a side view of the structure of a protein composed of two smaller proteins, called E and M. Each E and M contributes two molecules to the overall protein structure (called a heterotetramer), which is important for assembling and holding together the viral membrane, i.e., the shell that surrounds the genetic material of the dengue virus. The dengue protein's structure has revealed some portions in the protein that might be good targets for developing medications that could be used to combat dengue virus infections. For more on cryo-EM see the blog post Cryo-Electron Microscopy Reveals Molecules in Ever Greater Detail. You can watch a rotating view of the dengue virus surface structure in video 3748.
Hong Zhou, UCLA
View Media

3596: Heart rates time series image
3596: Heart rates time series image
These time series show the heart rates of four different individuals. Automakers use steel scraps to build cars, construction companies repurpose tires to lay running tracks, and now scientists are reusing previously discarded medical data to better understand our complex physiology. Through a website called PhysioNet developed in part by Beth Israel Deaconess Medical Center cardiologist Ary Goldberger, scientists can access complete physiologic recordings, such as heart rate, respiration, brain activity and gait. They then can use free software to analyze the data and find patterns in it. The patterns could ultimately help health care professionals diagnose and treat health conditions like congestive heart failure, sleeping disorders, epilepsy and walking problems. PhysioNet is supported by NIH's National Institute of Biomedical Imaging and Bioengineering as well as by NIGMS.
Madalena Costa and Ary Goldberger, Beth Israel Deaconess Medical Center
View Media

6780: Calling Cards in a mouse brain
6780: Calling Cards in a mouse brain
The green spots in this mouse brain are cells labeled with Calling Cards, a technology that records molecular events in brain cells as they mature. Understanding these processes during healthy development can guide further research into what goes wrong in cases of neuropsychiatric disorders. Also fluorescently labeled in this image are neurons (red) and nuclei (blue). Calling Cards and its application are described in the Cell paper “Self-Reporting Transposons Enable Simultaneous Readout of Gene Expression and Transcription Factor Binding in Single Cells” by Moudgil et al.; and the Proceedings of the National Academy of Sciences paper “A viral toolkit for recording transcription factor–DNA interactions in live mouse tissues” by Cammack et al. The technology was also featured in the NIH Director’s Blog post The Amazing Brain: Tracking Molecular Events with Calling Cards.
Related to video
Related to video
Allen Yen, Lab of Joseph Dougherty, Washington University School of Medicine in St. Louis.
View Media

2385: Heat shock protein complex from Methanococcus jannaschii
2385: Heat shock protein complex from Methanococcus jannaschii
Model based on X-ray crystallography of the structure of a small heat shock protein complex from the bacteria, Methanococcus jannaschii. Methanococcus jannaschii is an organism that lives at near boiling temperature, and this protein complex helps it cope with the stress of high temperature. Similar complexes are produced in human cells when they are "stressed" by events such as burns, heart attacks, or strokes. The complexes help cells recover from the stressful event.
Berkeley Structural Genomics Center, PSI-1
View Media

2513: Life of an AIDS virus
2513: Life of an AIDS virus
HIV is a retrovirus, a type of virus that carries its genetic material not as DNA but as RNA. Long before anyone had heard of HIV, researchers in labs all over the world studied retroviruses, tracing out their life cycle and identifying the key proteins the viruses use to infect cells. When HIV was identified as a retrovirus, these studies gave AIDS researchers an immediate jump-start. The previously identified viral proteins became initial drug targets. See images 2514 and 2515 for labeled versions of this illustration. Featured in The Structures of Life.
Crabtree + Company
View Media

6608: Cryo-ET cross-section of a rat pancreas cell
6608: Cryo-ET cross-section of a rat pancreas cell
On the left, a cross-section slice of a rat pancreas cell captured using cryo-electron tomography (cryo-ET). On the right, a 3D, color-coded version of the image highlighting cell structures. Visible features include microtubules (neon-green rods), ribosomes (small yellow circles), and vesicles (dark-blue circles). These features are surrounded by the partially visible endoplasmic reticulum (light blue). The black line at the bottom right of the left image represents 200 nm. Related to image 6607.
Xianjun Zhang, University of Southern California.
View Media

6748: Human retinal organoid
6748: Human retinal organoid
A replica of a human retina grown from stem cells. It shows rod photoreceptors (nerve cells responsible for dark vision) in green and red/green cones (nerve cells responsible for red and green color vision) in red. The cell nuclei are stained blue. This image was captured using a confocal microscope.
Kevin Eliceiri, University of Wisconsin-Madison.
View Media
2793: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 04
2793: Anti-tumor drug ecteinascidin 743 (ET-743) with hydrogens 04
Ecteinascidin 743 (ET-743, brand name Yondelis), was discovered and isolated from a sea squirt, Ecteinascidia turbinata, by NIGMS grantee Kenneth Rinehart at the University of Illinois. It was synthesized by NIGMS grantees E.J. Corey and later by Samuel Danishefsky. Multiple versions of this structure are available as entries 2790-2797.
Timothy Jamison, Massachusetts Institute of Technology
View Media
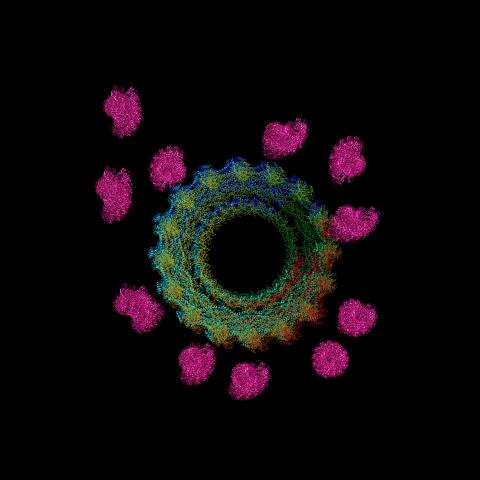
6571: Actin filaments bundled around the dynamin helical polymer
6571: Actin filaments bundled around the dynamin helical polymer
Multiple actin filaments (magenta) are organized around a dynamin helical polymer (rainbow colored) in this model derived from cryo-electron tomography. By bundling actin, dynamin increases the strength of a cell’s skeleton and plays a role in cell-cell fusion, a process involved in conception, development, and regeneration.
Elizabeth Chen, University of Texas Southwestern Medical Center.
View Media
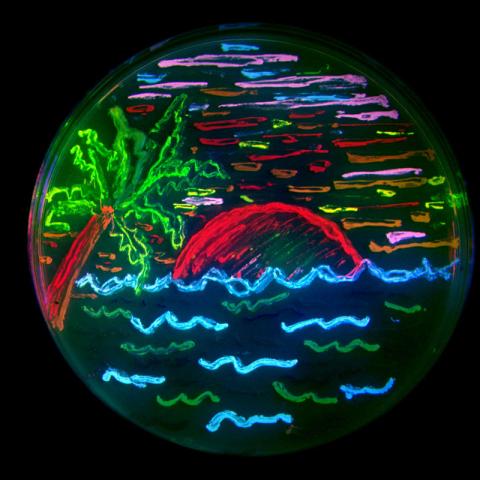
3492: Glowing bacteria make a pretty postcard
3492: Glowing bacteria make a pretty postcard
This tropical scene, reminiscent of a postcard from Key West, is actually a petri dish containing an artistic arrangement of genetically engineered bacteria. The image showcases eight of the fluorescent proteins created in the laboratory of the late Roger Y. Tsien, a cell biologist at the University of California, San Diego. Tsien, along with Osamu Shimomura of the Marine Biology Laboratory and Martin Chalfie of Columbia University, share the 2008 Nobel Prize in chemistry for their work on green fluorescent protein-a naturally glowing molecule from jellyfish that has become a powerful tool for studying molecules inside living cells.
Nathan C. Shaner, The Scintillon Institute
View Media

2572: VDAC video 03
2572: VDAC video 03
This video shows the structure of the pore-forming protein VDAC-1 from humans. This molecule mediates the flow of products needed for metabolism--in particular the export of ATP--across the outer membrane of mitochondria, the power plants for eukaryotic cells. VDAC-1 is involved in metabolism and the self-destruction of cells--two biological processes central to health.
Related to videos 2570 and 2571.
Related to videos 2570 and 2571.
Gerhard Wagner, Harvard Medical School
View Media

1336: Life in balance
1336: Life in balance
Mitosis creates cells, and apoptosis kills them. The processes often work together to keep us healthy.
Judith Stoffer
View Media

5855: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 1
5855: Dense tubular matrices in the peripheral endoplasmic reticulum (ER) 1
Superresolution microscopy work on endoplasmic reticulum (ER) in the peripheral areas of the cell showing details of the structure and arrangement in a complex web of tubes. The ER is a continuous membrane that extends like a net from the envelope of the nucleus outward to the cell membrane. The ER plays several roles within the cell, such as in protein and lipid synthesis and transport of materials between organelles. The ER has a flexible structure to allow it to accomplish these tasks by changing shape as conditions in the cell change. Shown here an image created by super-resolution microscopy of the ER in the peripheral areas of the cell showing details of the structure and the arrangements in a complex web of tubes. Related to images 5856 and 5857.
Jennifer Lippincott-Schwartz, Howard Hughes Medical Institute Janelia Research Campus, Virginia
View Media

3720: Cas4 nuclease protein structure
3720: Cas4 nuclease protein structure
This wreath represents the molecular structure of a protein, Cas4, which is part of a system, known as CRISPR, that bacteria use to protect themselves against viral invaders. The green ribbons show the protein's structure, and the red balls show the location of iron and sulfur molecules important for the protein's function. Scientists harnessed Cas9, a different protein in the bacterial CRISPR system, to create a gene-editing tool known as CRISPR-Cas9. Using this tool, researchers are able to study a range of cellular processes and human diseases more easily, cheaply and precisely. In December, 2015, Science magazine recognized the CRISPR-Cas9 gene-editing tool as the "breakthrough of the year." Read more about Cas4 in the December 2015 Biomedical Beat post A Holiday-Themed Image Collection.
Fred Dyda, NIDDK
View Media

2398: RNase A (1)
2398: RNase A (1)
A crystal of RNase A protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

3273: Heart muscle with reprogrammed skin cells
3273: Heart muscle with reprogrammed skin cells
Skins cells were reprogrammed into heart muscle cells. The cells highlighted in green are remaining skin cells. Red indicates a protein that is unique to heart muscle. The technique used to reprogram the skin cells into heart cells could one day be used to mend heart muscle damaged by disease or heart attack. Image and caption information courtesy of the California Institute for Regenerative Medicine.
Deepak Srivastava, Gladstone Institute of Cardiovascular Disease, via CIRM
View Media
1081: Natcher Building 01
1081: Natcher Building 01
NIGMS staff are located in the Natcher Building on the NIH campus.
Alisa Machalek, National Institute of General Medical Sciences
View Media

1272: Cytoskeleton
1272: Cytoskeleton
The three fibers of the cytoskeleton--microtubules in blue, intermediate filaments in red, and actin in green--play countless roles in the cell.
Judith Stoffer
View Media

3255: Centromeres on human chromosomes
3255: Centromeres on human chromosomes
Human metaphase chromosomes are visible with fluorescence in vitro hybridization (FISH). Centromeric alpha satellite DNA (green) are found in the heterochromatin at each centromere. Immunofluorescence with CENP-A (red) shows the centromere-specific histone H3 variant that specifies the kinetochore.
Peter Warburton, Mount Sinai School of Medicine
View Media

2408: Bovine trypsin
2408: Bovine trypsin
A crystal of bovine trypsin protein created for X-ray crystallography, which can reveal detailed, three-dimensional protein structures.
Alex McPherson, University of California, Irvine
View Media

3565: Podocytes from a chronically diseased kidney
3565: Podocytes from a chronically diseased kidney
This scanning electron microscope (SEM) image shows podocytes--cells in the kidney that play a vital role in filtering waste from the bloodstream--from a patient with chronic kidney disease. This image first appeared in Princeton Journal Watch on October 4, 2013.
Olga Troyanskaya, Princeton University and Matthias Kretzler, University of Michigan
View Media